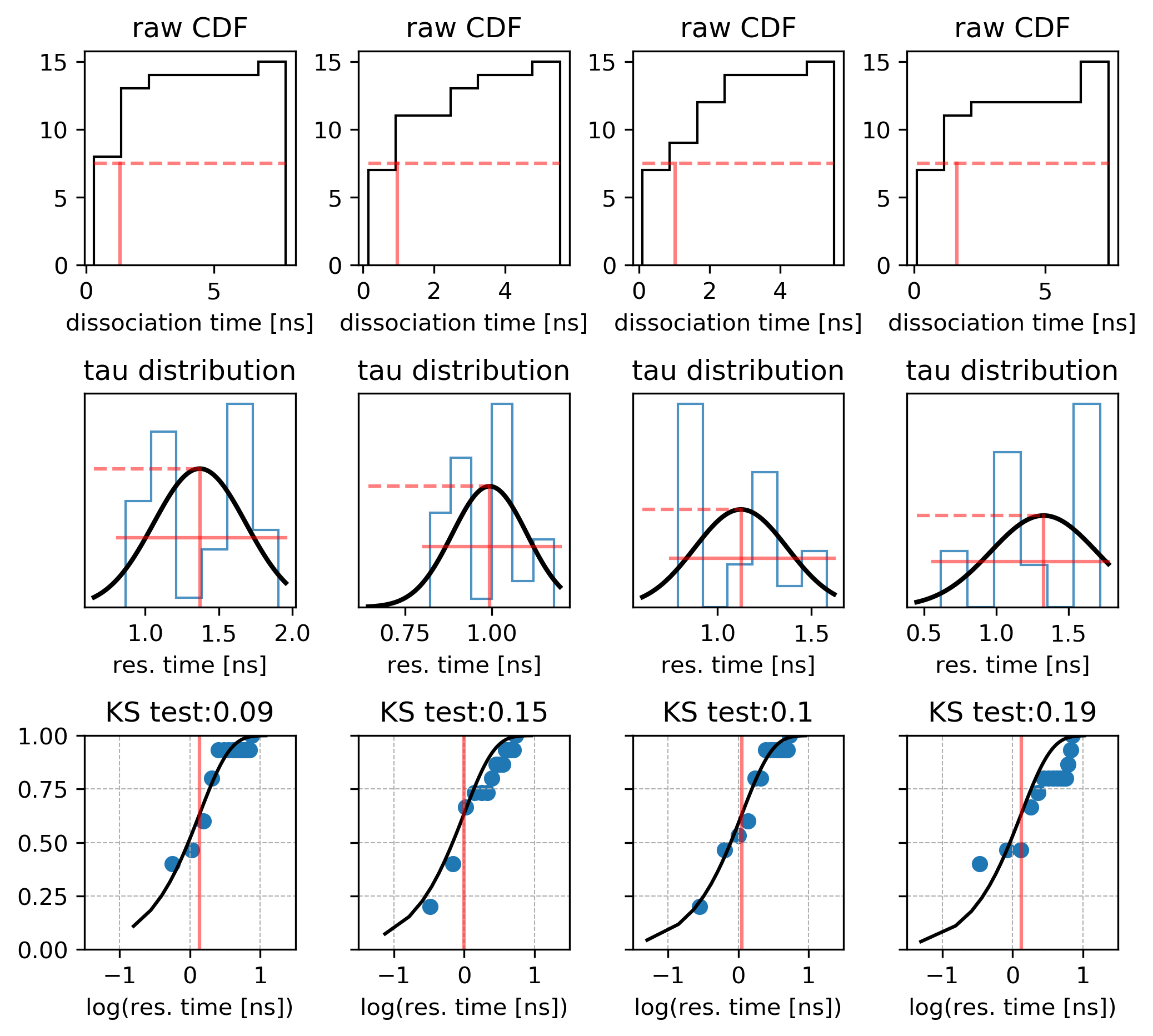
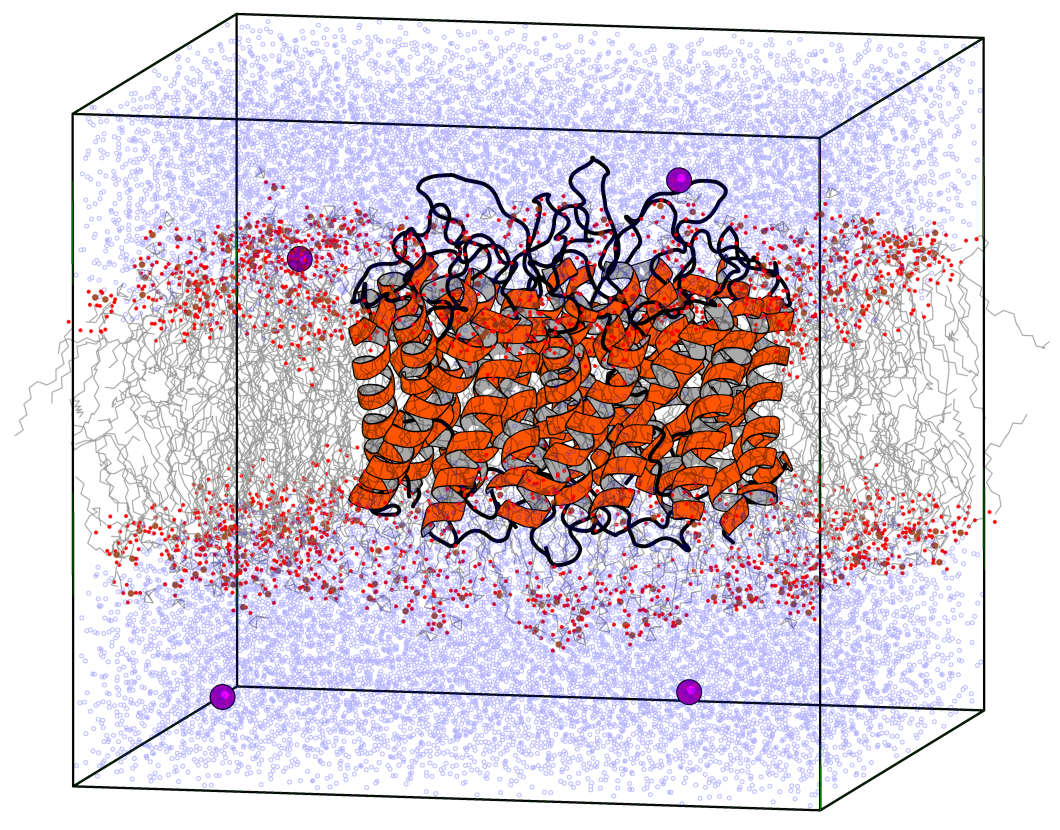
Using GPUs with Molecular Dynamics codes: optimizing usage from a user perspective Dr. Ole Juul Andersen CCK-11 February 1, ppt download

More bang for your buck: Improved use of GPU nodes for GROMACS 2018 - Kutzner - 2019 - Journal of Computational Chemistry - Wiley Online Library